Why: The migration from SAS to R aims to eliminate tool switching in PK/PD workflows, enable advanced analytics, and create a unified, future-ready environment for data analysis and AI integration.
What: The migration targets a SAS codebase used to generate analysis-ready datasets for a range of pharmacological and bioinformatic workflows including NCA, popPK, PBPK, and gene expression analyses.
Where: The migrated R code will be implemented using a combination of R Base, tidyverse, and pharmaverse packages - selected based on the specific needs of each analytical domain to balance compatibility with existing SAS logic, modern data handling, and CDISC-compliant workflows.
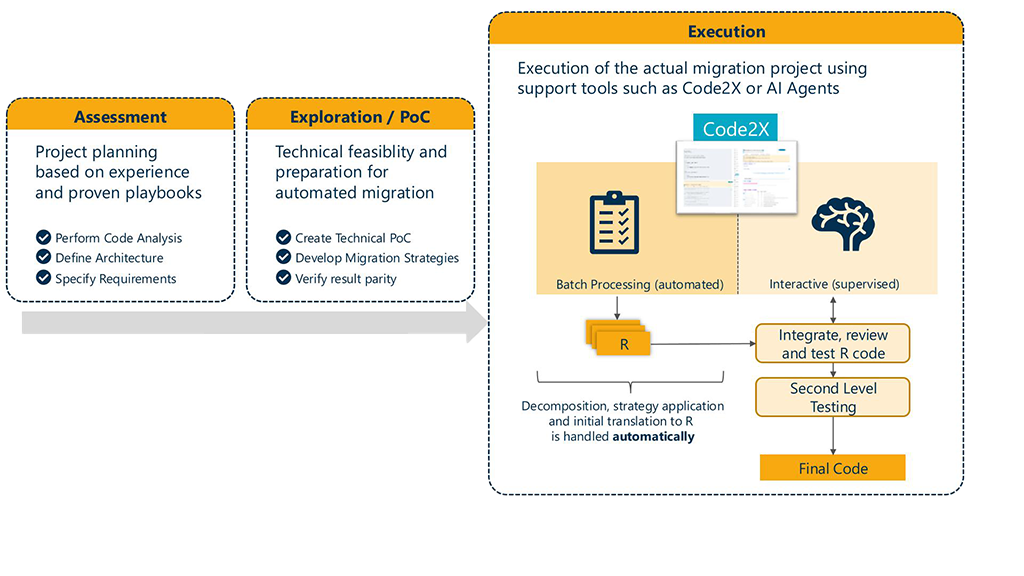
How: We started with an exploratory setup where we used the Ellmer R Package für LLM usage and structured output inside Posit Workbench Pro and extended this setup over time with automation.







